Evaluating and Improving SSU rRNA PCR Primer Coverage via Metagenomes from Global Ocean Surveys | bioRxiv
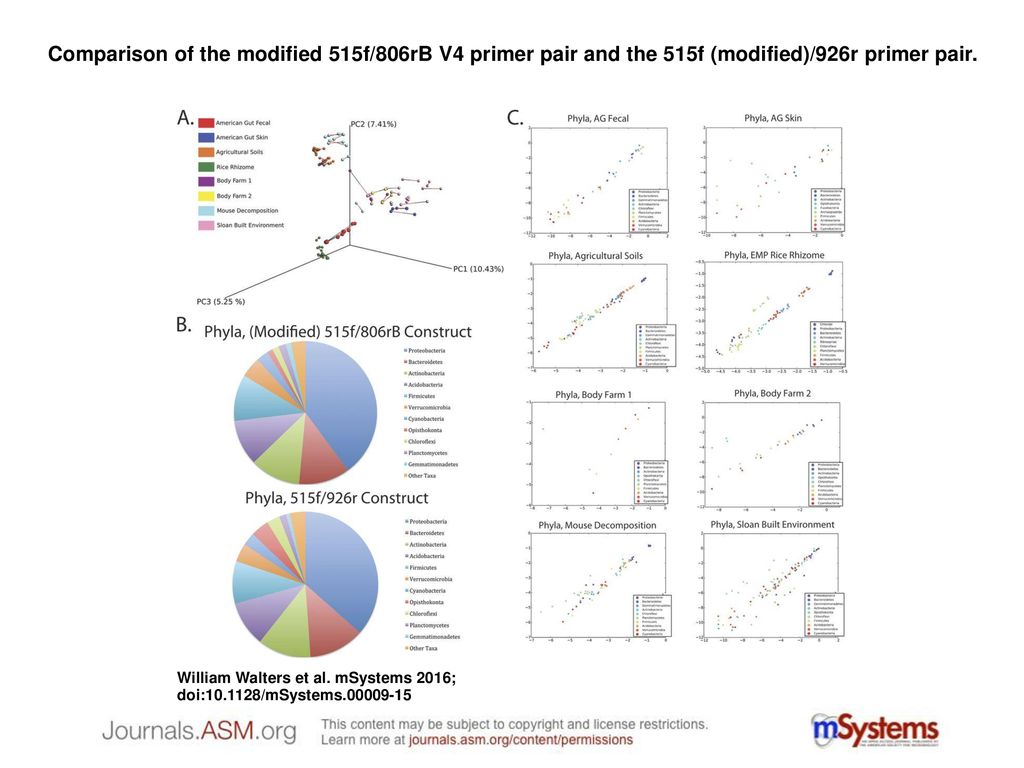
Comparison of the modified 515f/806rB V4 primer pair and the 515f (modified)/926r primer pair. Comparison of the modified 515f/806rB V4 primer pair and. - ppt download
The mean rarefied richness (number of ASVs, left) and Shannon Index (H... | Download Scientific Diagram

Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys | mSystems

Evaluating and Improving Small Subunit rRNA PCR Primer Coverage for Bacteria, Archaea, and Eukaryotes Using Metagenomes from Global Ocean Surveys | mSystems

Evaluating and Improving SSU rRNA PCR Primer Coverage via Metagenomes from Global Ocean Surveys | bioRxiv

Evaluating and Improving SSU rRNA PCR Primer Coverage via Metagenomes from Global Ocean Surveys | bioRxiv
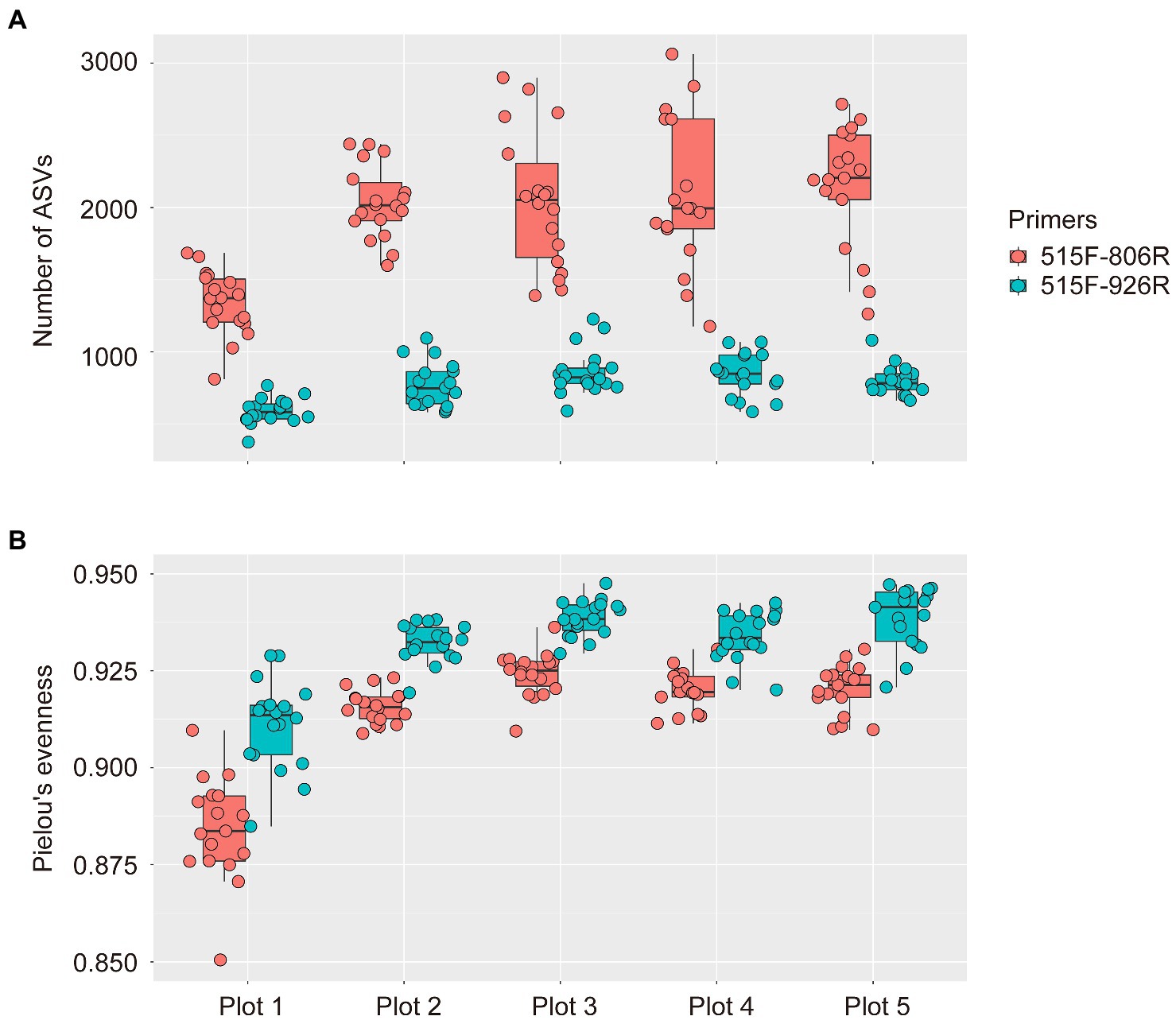
Frontiers | Fine-scale evaluation of two standard 16S rRNA gene amplicon primer pairs for analysis of total prokaryotes and archaeal nitrifiers in differently managed soils
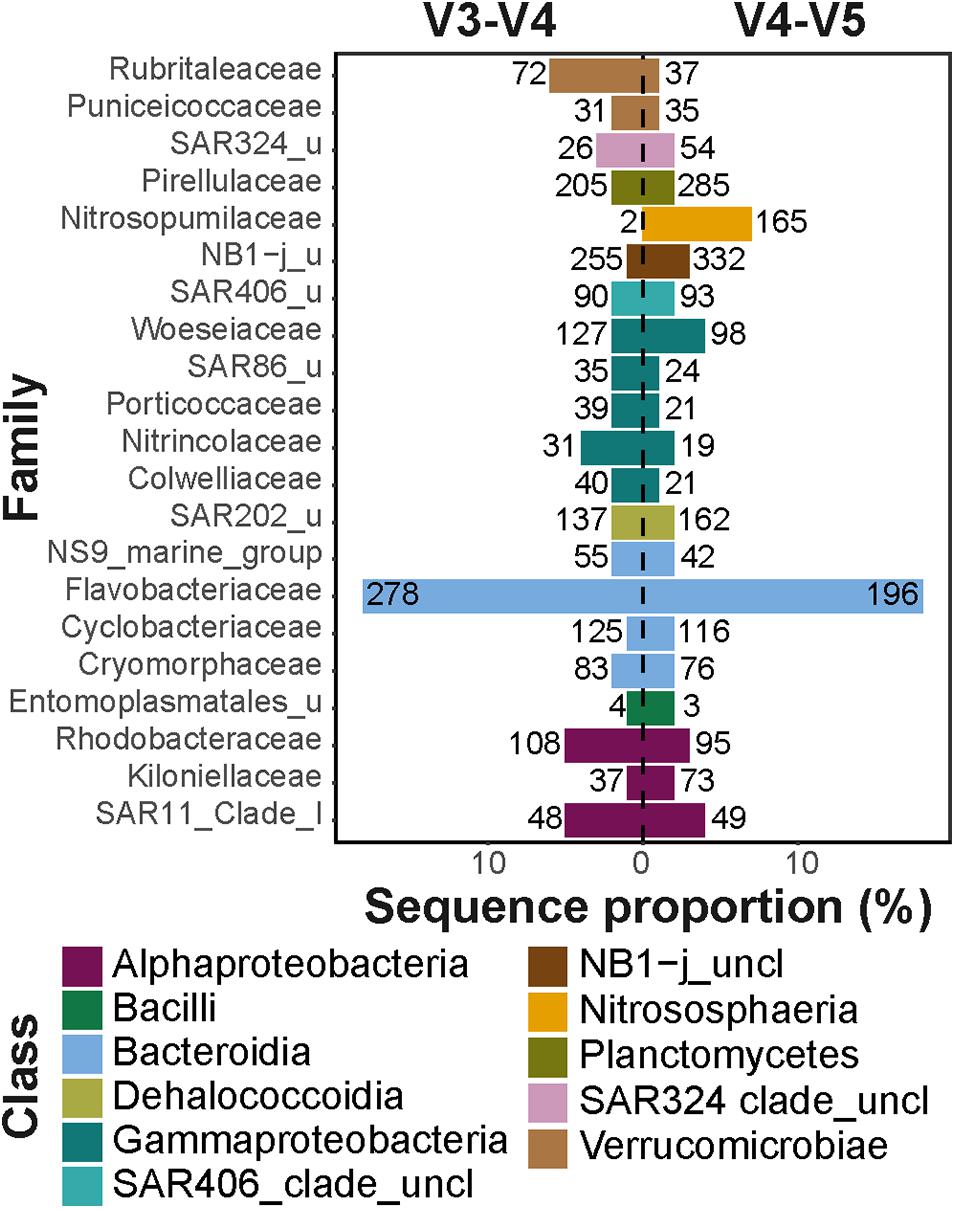
Frontiers | Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities

Specific and effective detection of anammox bacteria using PCR primers targeting the 16S rRNA gene and functional genes - ScienceDirect
Supplemental Digital Content METHODS 16S rRNA Amplification For each fecal specimen, duplicate 50-μL PCR reactions were perform

Evaluating and Improving Small Subunit rRNA PCR Primer Coverage for Bacteria, Archaea, and Eukaryotes Using Metagenomes from Global Ocean Surveys | mSystems
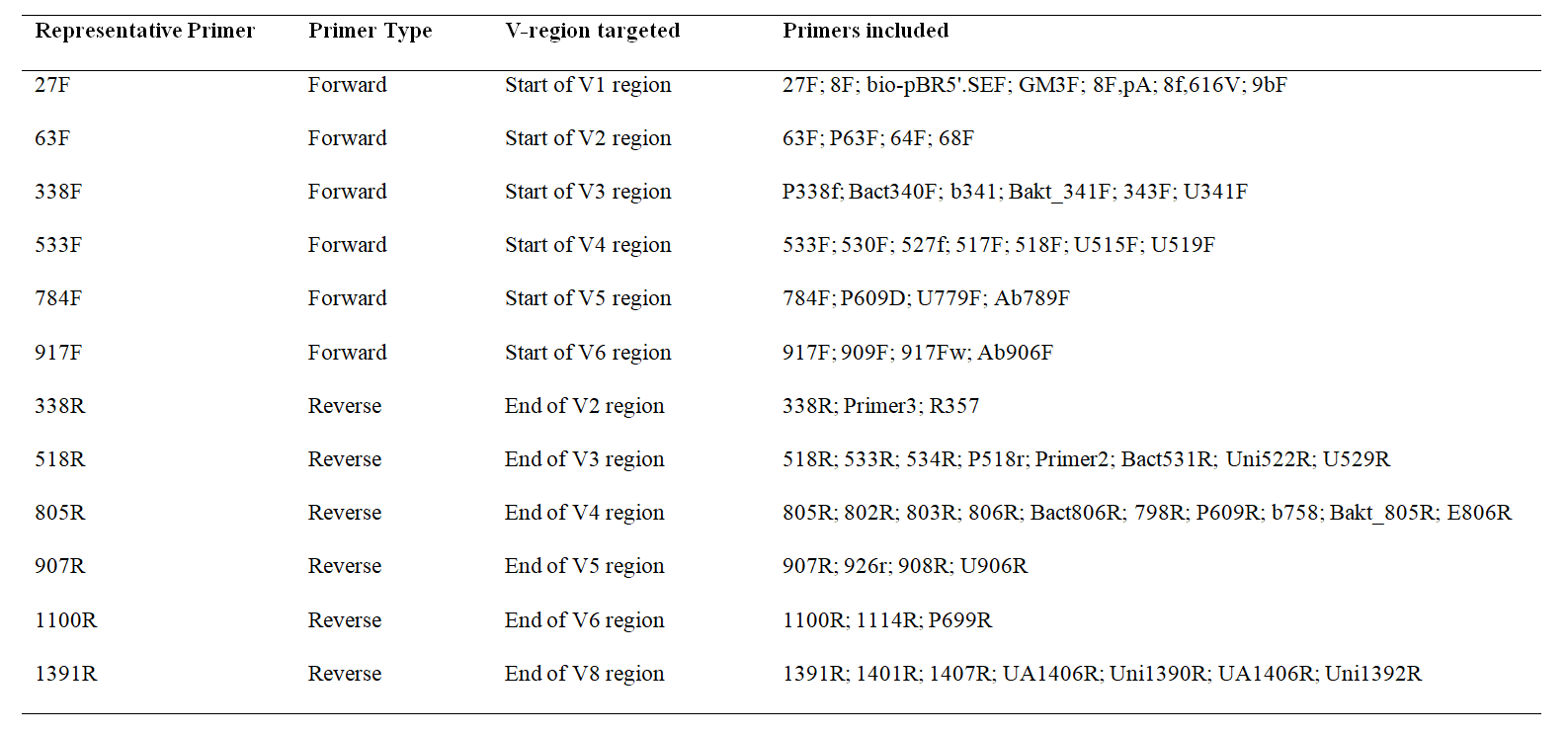
![PDF] Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. | Semantic Scholar PDF] Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/fca711f0e64e829a43ed532f0f1a3fb0c47e0e9d/4-Table1-1.png)
PDF] Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. | Semantic Scholar