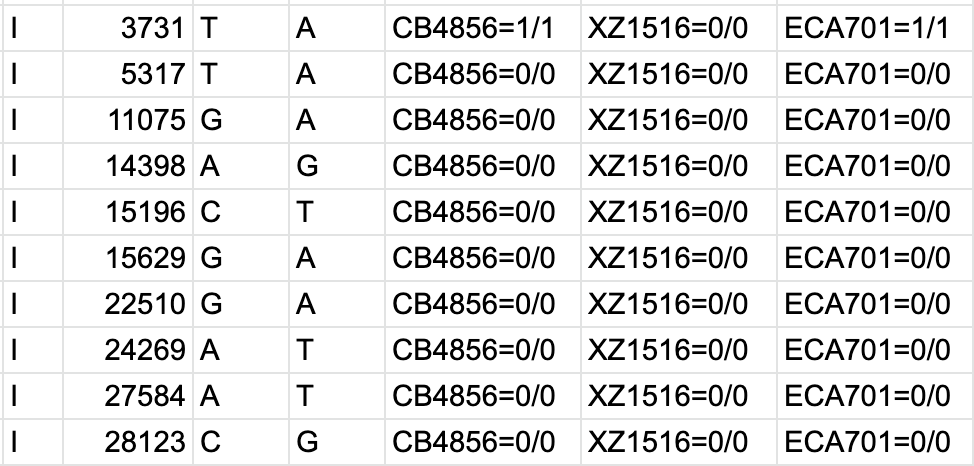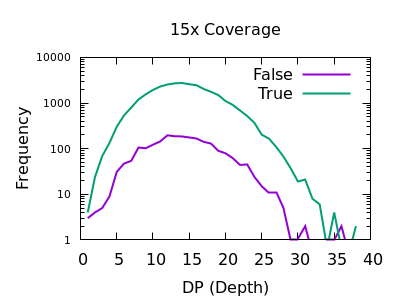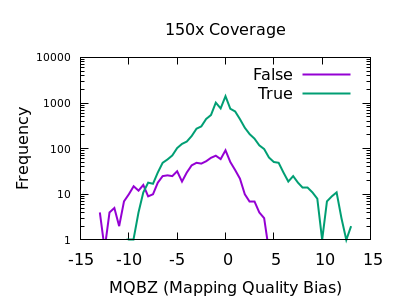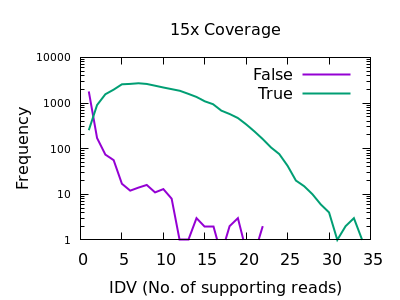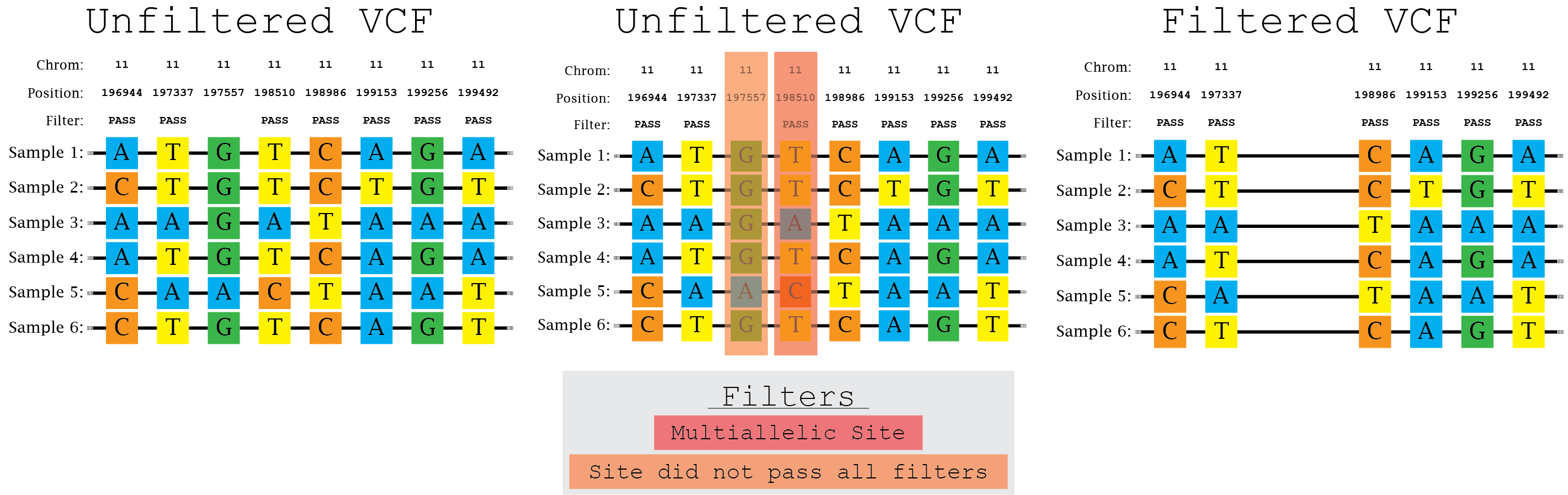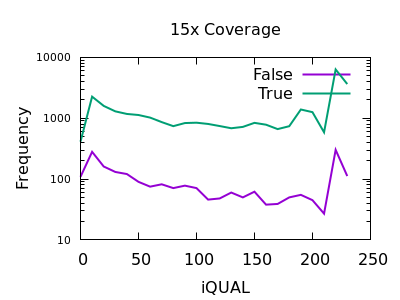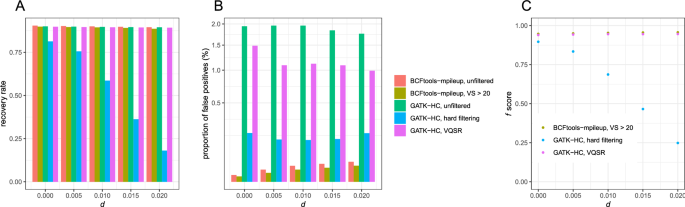
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports

bcftools mpileup - Difference between IDV and FORMAT/AD* fields · Issue #912 · samtools/bcftools · GitHub

Cells | Free Full-Text | A Next Generation Sequencing-Based Protocol for Screening of Variants of Concern in Autism Spectrum Disorder
Comparing -min-DP in vcftools with filter -i 'FORMAT/DP>10' in bcftools · Issue #1384 · samtools/bcftools · GitHub
Feature request] Filter BCF/VCF on genotype state and count for the minor allele · Issue #544 · samtools/bcftools · GitHub
Possible to annotate the FILTER from an annotation VCF file, as an INFO/TAG in the target? · Issue #1187 · samtools/bcftools · GitHub